
Included in the publication is the profiling of a range of small molecule compounds and biological stimuli which demonstrated the potential of the method to identify and study membrane – bound protein targets. , CETSA ® Explore was combined with selective enrichment of glycosylated cell surface proteins. Alternating amphiphilic multiblock molecules 14, involving fluorescent hydrophobic units, were designed as mimics for multipass transmembrane proteins. This is hugely valuable to drug discovery researchers in that by demonstrating how CETSA ® can be used for membrane-bound proteins, such as ion channels and transporters, scientists now have an alternative to biochemical or biophysical methods. This stud y describe s the thorough experimental work undertaken to develop assay protocols for the more challenging multi-pass transmembrane proteins. approaches in both yeast and human cells revealed that the ER membrane protein complex (EMC) binds to and promotes the biogenesis of a range of multipass.
#Multipass transmembrane protein how to
Ĭase studies on how to deploy CETSA ® on insoluble protein targets such as membrane proteins have been published, for example by Kaw atkar et al. Table 1: Antigens for membrane protein antibody drug discovery Membrane Nanoparticles (MNPs) Plasma membrane-coated nanoparticles (MNPs) have been used in various applications, including delivery of therapeutic agents and induction of immune responses et al. Protein sets from fully sequenced genomes. Figure 2: DIMA’s solutions for the full-length multipass transmembrane proteins.

#Multipass transmembrane protein archive
Nevertheless, CETSA ® has successfully been applied to a number of membrane- associated proteins including GPCR s and transporter s. Help pages, FAQs, UniProtKB manual, documents, news archive and Biocuration projects. Recent years, increasing evidence has showed that transmembrane proteins (also known as multi-pass transmembrane proteins) are great potential targets of.
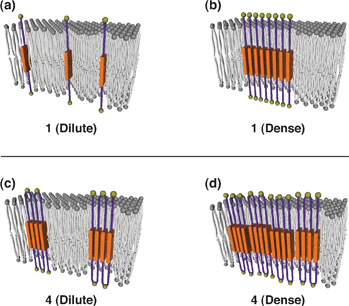
Our results show that there are characteristic preferences for residues to face the headgroup region and the hydrocarbon core region of lipid membrane. Therapeutic antibodies to this class of proteins are generally targeted to extracellular domains displayed on the surfaces of living cells. Lipid accessibilities of amino acid residues within the transmembrane (TM) region of 29 structures of helical membrane proteins are studied with a spherical probe of radius of 1.9 Å.
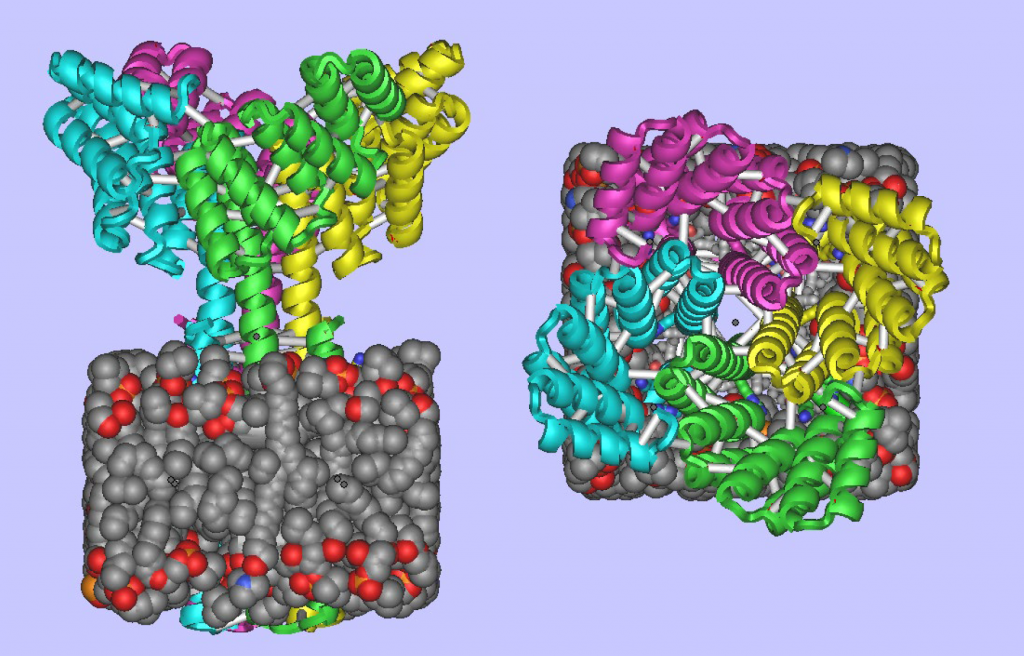
Transmembrane proteins, including multipass transmembrane proteins like GPCRs and ion channels, are important targets for therapeutic monoclonal antibody (mab) discovery. Accurate computational design of multipass transmembrane proteins.


 0 kommentar(er)
0 kommentar(er)
